What Is Drug Resistance?
Drug resistance occurs when microorganisms experience changes that make medications or disinfectants less effective. This can be caused by events like a bacterium independently developing resistant genes, a microbe with naturally occurring resistant genes being allowed to multiply, or bacteria sharing resistant genes among each other. When these events happen, existing drugs lose their effectiveness.
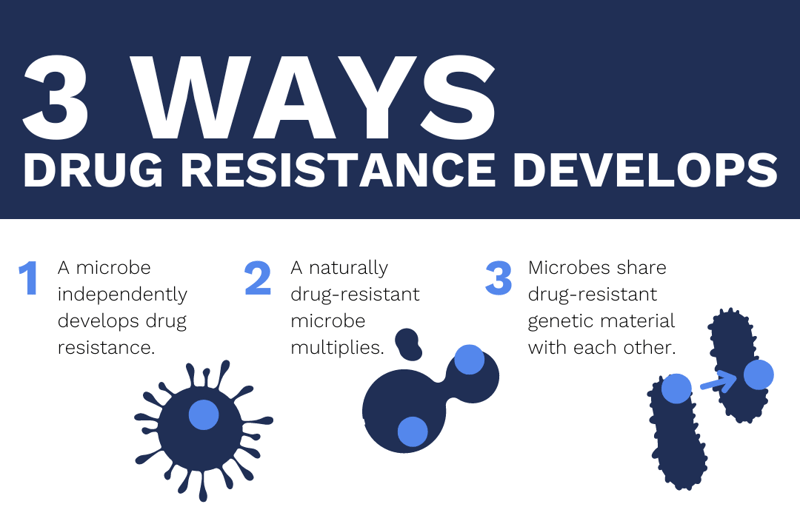
This article will focus on antimicrobial drug resistance, as well as disease and drug research.
In This Article
- How Is Microbial Resistance Tested?
- Benefits of WGS in Drug Resistance
- Examples of Whole Genome Sequencing in Drug-Resistance Testing
- WGS and the Future of Personalized Medicine
How Is Microbial Resistance Tested?
This article discusses three main methods of antimicrobial drug-resistance testing (DRT). The first method tests microbes through bacterial isolation, culture, and drug exposure. A microbe is isolated, then exposed to the drugs relevant to the study.
Depending on how many drugs are in consideration, this process can take weeks or months to complete. In a clinical setting, the risk of misdiagnosis is high when using this drug resistance screening method. It is ineffective when an infection is caused by a mix of isolates or by unrelated bacterial strains.
The second drug-resistance testing method is a Polymerase Chain Reaction (PCR) based test. PCR technology allows researchers to rapidly amplify specific gene sequences associated with drug resistance in a bacterial species.
In 2008, the World Health Organization (WHO) endorsed an option for testing drug-resistant tuberculosis: molecular line probe assay (LPA). This process is much faster than drug resistance screening.
In this method, PCR technology is used to amplify drug-resistant gene regions. Then, the PCR products are immobilized on a strip, and detection is achieved via a colorimetric reaction. While this is a faster test than isolation/exposure, PCR testing requires previous knowledge about which parts of the genome can cause drug resistance.
Whole-genome sequencing (WGS) is a more open-ended option. It allows researchers to investigate a microbe when they aren’t sure what part of its genome houses drug-resistant genetic material.
Benefits of WGS in Drug Resistance
The WGS approach to drug testing accesses previously unknown sequence information. This is a significant advantage to other methods that require researchers to know where to look. High-throughput WGS has improved power and resolution, and has already made advancements in identifying and tracking drug-resistant organisms.
Examples of Whole Genome Sequencing in Drug-Resistance Testing
Clinicians have already made use of WGS for DRT in health care settings. Below are some ways WGS has improved health outcomes, prevented additional infections, and provided insights to combat evolving microbes.
Infection detection and treatment
WGS has identified many genetic determinants of drug resistance in pathogenic microbes (such as ampC in E. coli and mecC in S. aureus). This has been applied in clinical settings when other diagnostic and therapeutic interventions fail.
In one case, a patient was admitted to the emergency room with a soft tissue infection and cardiomyopathy. Blood cultures revealed an infection by a β-lactamase producing E. coli. WGS performed on bacterial isolates uncovered several resistance genes, including ampC2. Because WGS was used, clinicians chose a treatment based on the irregularity of the infection, and the patient made a full recovery.
Infectious disease prevention
Whole-genome sequencing was used to investigate a suspected outbreak of methicillin-resistant S. aureus (MRSA) in a special care baby ward in Cambridge, UK. Infection control officials conducted antimicrobial susceptibility testing on MRSA samples collected from mothers and infants. They found evidence of repeat MRSA infections with the same genetic variant.
When a deep clean of the facility did not stop transmission, researchers theorized they were dealing with an asymptomatic carrier on their staff. A screening of hospital staff for MRSA found one carrier, who was removed from clinical duties until they could be treated and decolonized.
WGS in tuberculosis research
The public health threat from tuberculosis is particularly challenging because of the existence of multi-, extensively, and totally drug-resistant tuberculosis strains. Whole-genome sequencing technology allows for the complete characterization of drug resistance in tuberculosis samples.
Coll, et al. developed the TB-Profiler tool to bolster drug-resistance testing efforts. The online TB-Profiler tool inputs raw sequence data (from a library of over 1,000 mutations), identifies drug resistance and lineage-specific mutations, and outputs the corresponding data.
This analytical approach provides a means to predict resistance to the 11 anti-TB drugs tested. Given the need for early diagnosis of tuberculosis and subsequent effective and minimally or non-toxic treatment, having the full drug-resistance profile of strains would greatly improve treatment success.
Predicting resistant pathogens
Sequencing technology can do more than identify current drug-resistant pathogens — it can also provide clues to a pathogen’s potential to develop drug resistance. Proto-resistance genes are the precursors to resistant genes, and can tell researchers if a microbe has the potential to evolve drug resistance in the future. WGS is used to identify these genes, as well as other determinants of resistance.
These discoveries are cataloged in databases like the National Database of Antibiotic Resistant Organisms (NDARO), the Comprehensive Antibiotic Resistance Database (CARD), and the Antibiotic Resistant Target Seeker Database (ARTS-DB). The information in these databases has been used to develop assays and test kits for clinical laboratory work.
The databases also have great potential for use in developing new treatments. For years, concerns about new superbugs have warned of an increase in viral and bacterial resistance to drugs; understanding the genetic makeup of superbugs makes it possible for pharmaceutical researchers to predict and understand evolving microbes.
WGS and the Future of Personalized Medicine
For WGS to be a meaningful tool in detecting and combating antimicrobial drug resistance, the clinical space would need two key pieces of infrastructure. The first is easy-to-implement bioinformatics tools for clinicians and hospitals. For widespread adoption, health care professionals will need accessible technologies that provide clear results. The second is a series of validation protocols, ensuring that results can be interpreted with confidence.
Researchers anticipate that the adoption of WGS technology in drug-resistance research will result in:
- Personalized therapies. Clinicians will be able to treat a patient for their specific infectious disease. Accessible, routine DNA sequencing in health care will eliminate guesswork and prevent the use of unnecessary or ineffective antimicrobial treatments.
- Antibiotic development. As microbes change, so do the drugs used to fight them. Understanding the way microbes are changing around the world will make it easier to develop drugs that will be effective against them.
- Improved infectious disease recovery. In their 2019 Antibiotic Resistance Threats report, the Centers for Disease Control and Prevention estimated 2.8 million antibiotic-resistant infections occur in the United States every year, causing over 35,000 annual deaths. Having the tools necessary to combat microbial drug resistance will improve survival and recovery rates.
Whole-genome sequencing is just getting started with all it can contribute to drug research efforts! To learn more about applications of WGS, see our whole-genome sequencing page.
