Next Generation Sequencing vs. Sanger Sequencing
DNA sequencing uses biochemical processes to determine the order of nucleotides in DNA strands. Sanger sequencing was developed over 40 years ago, but is still a widely used tool in genomic research.
Although Sanger sequencing is commonly used, it can be expensive and time-consuming. In recent years, next-generation sequencing has overcome many limitations of Sanger sequencing. Massively parallel technologies are able to perform large-scale sequencing projects with improved speed and cost effectiveness.
This article explores how Sanger sequencing and NGS work, how NGS improves upon Sanger sequencing, and how the two technologies can be combined for major discoveries.
How Does Sanger Sequencing Work?
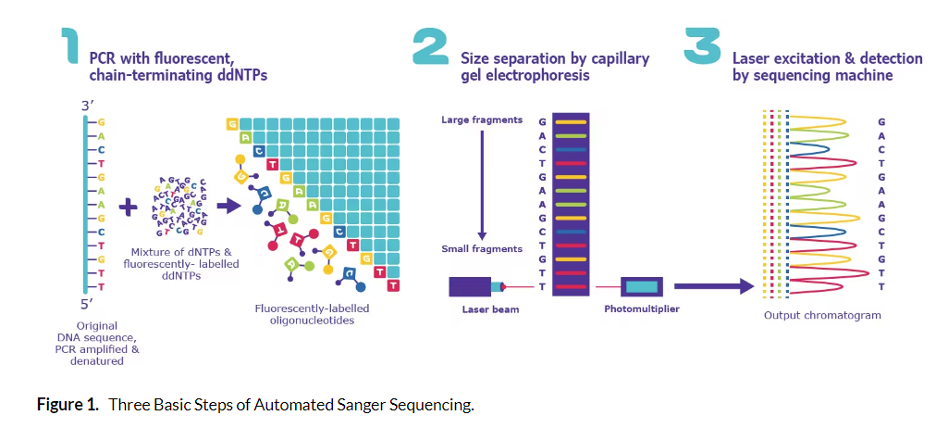
In Sanger sequencing, targeted regions of DNA are sequenced. Short single-stranded DNA oligonucleotides complementary to the region of interest are attached at short intervals across the DNA region of interest. Then, a polymerase extends each primer by
The nucleotide pools used to grow the new DNA chain contain a mixture of unlabeled bases and a fraction of chain terminator bases (G, A, T, C). In addition, the labels on the terminator bases each have a different color fluorescent tag so that as each region of the target DNA is copied, the terminator bases are added and the fluorescent signal is captured. When polymerase encounters the terminator dideoxynucleotides, it cannot add additional nucleotides to the sequence.
This generates fragments that “terminate” at every position in the sequence being copied. These fragments are separated by chromatography inside the sequencing instrument and the fluorescent terminator bases are used to produce a chromatogram of the full region being analyzed. Interestingly, when Nobel Laureate Frederick Sanger first invented DNA sequencing, fragments were actually run on gigantic gels.
Sanger sequencing has led to many important discoveries over the years and is currently the only FDA approved sequencing method for drug manufacturing. However, technological advancements have led to new approaches that surpass Sanger in terms of scale and affordability.
In recent years, next-generation sequencing has overcome the costly and time-consuming aspects of using Sanger sequencing on a wide/population-level scale. Massively parallel technologies are able to perform large-scale sequencing projects with improved speed and cost effectiveness ushering in the era of next generation sequencing (NGS).
The Next-Generation Sequencing Era
Next-generation sequencing is a massive parallel high-throughput sequencing technology. All NGS approaches are characterized by conducting millions of concurrent sequencing reactions (parallel reactions).
With the development of NGS technologies, genome sequencing is faster and more affordable. In the year 2000, it cost nearly $100 million to sequence a human genome using the Sanger method. With next-generation sequencing, a human genome can be sequenced for under $1000.
How Does NGS Work?
NGS processes are responsible for the speed and power delivered by today’s sequencing approaches. First, a library of the nucleic acid being analyzed is generated. If RNA is the sample, the RNA is converted to cDNA that is used to create the library. Next, the DNA strands are broken up and short adapters are ligated onto the end of every fragment.
The adapters can serve multiple functions, including binding to the flow cell, providing unique barcodes that permit sample pooling, unique molecular identifiers (UMIs) for data quality control and quantitation. Finally, the adapters serve as primer binding sites for amplification either preceding and/or during the sequencing reaction.
Libraries vary widely depending on the type of nucleic acid being analyzed, the information desired, and the specific sequencing instrument (NovaSeq X, PacBio Revio, T7, e.g.). Libraries are typically amplified before the actual sequencing phase to increase sensitivity. The library fragments are then bound to a flow cell by their adapters. Sequencing is often described as “sequencing by synthesis.”
Each library fragment bound to the flow cell is copied using labeled nucleotides that are sequentially imaged as they are incorporated into the growing DNA chain. The incorporation of the nucleotides is detected by various techniques depending on the sequencing instrument. For example, Illumina short-read sequencing uses different colors of fluorescently-labeled nucleotides to distinguish among the four DNA bases (G, A, T, C) as they are added to the growing fragment.
After all the fragments have been copied and their nucleotide contents captured (AKA their sequences), the data is processed to remove adapters and artifacts. The fragments are then assembled to a reference genome. Analyzing these high content data sets enables researchers to make amazing discoveries that we couldn’t have dreamed of before the invention of NGS.
NGS Advancing Research and Clinical Medicine
NGS has rapidly changed many medical and scientific fields. Personalized medicine, epidemiology, forensics, and basic scientific discovery have benefitted from inexpensive, rapid technologies. In rare and orphan diseases like Aspartylglucosaminuria (AGU), NGS has made diagnosis and development of targeted gene therapies a possibility.
NGS can generate accurate, high-throughput analysis of the genome, transcriptome, and proteome. Some instruments, like the 10x Genomics Xenium, even provide the subcellular coordinates of each transcript within every individual cell in a tissue. These capabilities provide different perspectives and insights, creating value in disease research and exploratory efforts.
In the years since NGS was introduced, large reference data sets have led to improvements in diagnostic procedures. Having sequencing information readily available makes it possible to develop targeted laboratory tests that can be validated and utilized in a clinical setting.
For example, genetic disorders are a leading cause of illness and death for infants in neonatal wards. Although additional research is required, a 2018 trial had success using rapid whole genome sequencing (rWGS) to diagnose infants with unknown illnesses.
rWGS provided a diagnosis at an average of 25 days from enrollment in the study. In comparison, control group infants averaged 130 days between enrollment and diagnosis. These are promising results for diseases that require a timely diagnosis.
Joining Forces: NGS + Sanger Sequencing
Sanger sequencing is still an important tool in genomic research. Often, Sanger is used to verify NGS sequencing data. This is particularly useful when NGS has uncovered new information in basic research efforts. Often, Sanger sequencing is used to confirm the accuracy of these results.
This is helpful when researchers are concerned about false positive or negative results. Although NGS continues to improve in speed and accuracy, Sanger remains the gold standard for validating results in diagnostic settings.
Rapid, cost-effective NGS technologies are changing sequencing capabilities in many settings. In clinical settings, discoveries from NGS have led to major expansions in pharmacogenomic testing, alleviating treatment risks for a number of medications whose side effects could be life-threatening, albeit for a subset of patients. This improves diagnostic capabilities that were previously cost-prohibitive and slow.
Although Sanger sequencing remains an essential tool in genetic research and medicine, NGS has made many things possible that would previously have been impossible due to the long, costly process of Sanger sequencing large numbers of participants.
Psomagen thanks Dr. Stacy Matthews Branch for her contributions to the research and writing of the original version of this article. Dr. Branch is a biomedical consultant, medical writer, and veterinary medical doctor. She owns Djehuty Biomed Consulting and has published research articles and book chapters in the areas of molecular, developmental, reproductive, forensic, and clinical toxicology. Dr. Matthews Branch received her DVM from Tuskegee University and her Ph.D. from North Carolina State University.